Protein Synthesis In Eukaryotes Definition
During protein synthesis, the messenger RNA (mRNA) molecule is translated into the sequence of amino acids. The mRNA is produced following the transcription process of DNA which is utilized by the cytoplasmic or endoplasmic reticulum (ER) ribosomes create to produce a stretch of a polypeptide or protein.

The Ribosomes
- Typically, ribosomes comprise separate subunits composed of protein as well as rRNA.
- Eukaryotic ribosomes are more complex in nature and bigger (the 80S) than prokaryotic ribosomes (the 70S).
- When the ribosomal subunits attach to an mRNA near its 5′ end, they assemble into a ribosome.
- The ribosome interprets the 5′ to 3′ nucleotide sequence upon attaching to an mRNA, producing the appropriate protein from amino acids in an N-terminal (amino-terminal) to C-terminal (carboxyl-terminal) manner.
- In the cytoplasm, ribosomes are either free-floating or linked with the endoplasmic reticulum.
- They are necessary for protein synthesis.
Ribosomal Sites for Protein Synthesis
Schematically, every ribosome possesses three tRNA binding sites.
- The aminoacyl-tRNA binding site (or A site) is where aminoacyl-tRNA interacts with incoming aminoacyl-tRNA during elongation.
- The peptidyl-tRNA binding site (or P site) is where the tRNA is linked to the growing polypeptide chain bonds.
- The exit site (or E site) is a tRNA-binding site prior to its release from the ribosome following its function in translation.
All three ribosomal sites (A, P, and E) are produced by rRNA molecules.
Protein Synthesis Process
Comparatively, the methods of protein production in eukaryotes and prokaryotes are essentially the same.
However, there are important distinctions:
- A bacterial ribosome has a 70S sedimentation coefficient and 30S and 50S subunits, whereas a eukaryotic ribosome has an 80S sedimentation coefficient and 40S and 60S subunits.
- Similarly, the constituents of eukaryotic ribosomal subunits are more complicated than those of bacterial subunits, yet the activity of every subunit is largely identical to that of prokaryotic subunits.
- In eukaryotes, every mRNA is monocistronic, encoding a single protein, excluding any potential post-translational cleavage processes. Many mRNAs in prokaryotes are polycistronic, encoding certain proteins. Each codon in a bacterial mRNA coding sequence possesses its own start and stop codons.
- In contrast to prokaryotes, the commencement of protein synthesis in eukaryotes needs at least nine unique eukaryotic initiation factors (eIFs). Prokaryotes only require three initiation factors (IFs).
- In eukaryotes, the initiating amino acid is methionine, whereas in prokaryotes it is N-formylmethionine.
- As in prokaryotes, initiation requires a specific initiator tRNA separate from the tRNA that identifies and binds to methionine codons at internal locations in the mRNA. This is termed as Met-tRNAimet when it is loaded with methionine and ready for initiation.
- In prokaryotes, a Shine–Dalgarno sequence 5′ to the AUG initiation codon serves as the binding site for the 30S ribosomal subunit, but in eukaryotes, the 30S ribosomal subunit binds to the AUG initiation codon.
- In contrast, the majority of eukaryotic mRNAs lack Shine–Dalgarno sequences. Instead, a 40S ribosomal subunit binds to the 5′ end of the mRNA and travels downstream (i.e., from 5′ to 3′) until it reaches the AUG start codon. This method is known as scanning.
- Prokaryotic translation does not require helicase, presumably because protein synthesis in bacteria can begin while the mRNA is still being synthesized, whereas, in eukaryotes, transcription in the nucleus and translation in the cytoplasm to occur independently, allowing time for the formation of mRNA secondary structure.
You may also like to read: Vacuoles
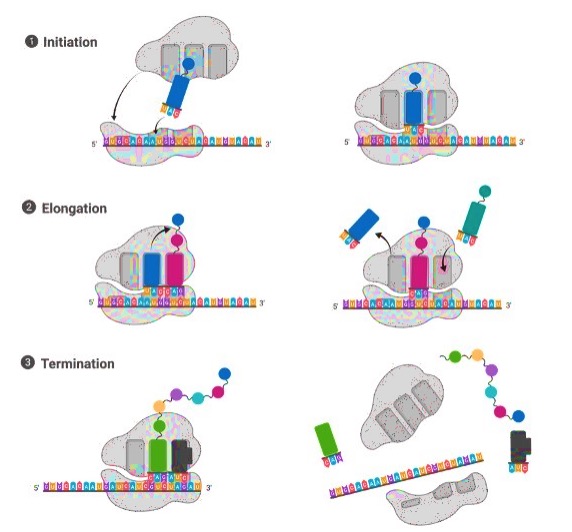
The three phases of protein synthesis (or translation) are as follows:
1) Initiation
2) Elongation and
3) Termination
Initiation of Protein Synthesis
- The creation of a pre-initiation complex made up of the 40S small ribosomal subunit, Met-tRNAimet, eIF-2, and GTP is the initial step.
- eIF-4F, also known as the cap-binding complex, and eIF-3 are needed for the pre-initiation complex to attach to the 5′ end of the eukaryotic mRNA.
- The three components of the eIF-4F complex, eIF-4A, eIF-4E, and eIF-4G, engage with the poly (A) binding protein on the poly (A) tail while eIF-4E attaches to the 5′ cap of the mRNA.
- An ATP-dependent RNA helicase called eIF-4A unravels any secondary structures in the mRNA to make it ready for translation.
- The complex then proceeds in a 5′ to 3′ orientation along the mRNA until it finds the AUG initiation codon (i.e., scanning).
- The 5′ untranslated region of a eukaryotic mRNA can contain hundreds of nucleotides and may contain secondary structures like hairpin loops. These secondary structures are likely eliminated by the scanning complex’s initiation elements.
- The Kozak consensus (5′-ACCAUGG-3′) is a brief sequence that frequently (but not always) contains the initiation codon, which makes it easy to identify.
- The 60S large ribosomal subunit binds to create an 80S initiation complex after the complex is positioned over the start codon. This action necessitates the hydrolysis of GTP and results in the release of a number of initiation factors.
Elongation of Protein Synthesis
- Eukaryotic elongation factors are necessary for elongation.
- eEF-1A, eEF-IB, and eEF-2, three elongation factors with comparable roles to their prokaryotic counterparts, EF-Tu, EF-Ts, and EF-G, are involved.
- In order for the subsequent codon to be translated during the elongation phase of protein synthesis, the mRNA is placed at the end of the initiation step.
- The initiator tRNA occupies the P site of the ribosome, whereas the A site is open for an aminoacyl-tRNA.
- In a three-step microcycle, each additional amino acid is added to the growing polypeptide chain during chain elongation.
The steps in this microcycle are:
- Placing the appropriate aminoacyl-tRNA in the ribosome’s A site,
- The peptide bond is created and
- Shifting the mRNA in relation to the ribosome by one codon.
- The mRNAs created within the organelles of certain eukaryotes employ a different genetic code, despite the fact that most codons in prokaryotes and eukaryotes encode the same amino acids.
- The deacylated tRNA in the P site transfers to the E site before exiting the ribosome during elongation in bacteria. The deacylated tRNA appears to be expelled straight from the ribosome in eukaryotes, despite the fact that the situation is still not entirely apparent.
Termination of Protein Synthesis
- Eukaryotic release factors are necessary for the termination of elongation.
- Eukaryotic release factor eRF-1, which is found in eukaryotes, recognizes all three termination codons (UAA, UAG, and UGA) and together with the protein eRF-3, stops translation.
- When the process is finished, the ribosome is broken down, and the finished polypeptide is released.
References
- Protein Synthesis: Definition, Steps, and Diagram
- Eukaryotic Protein Synthesis





